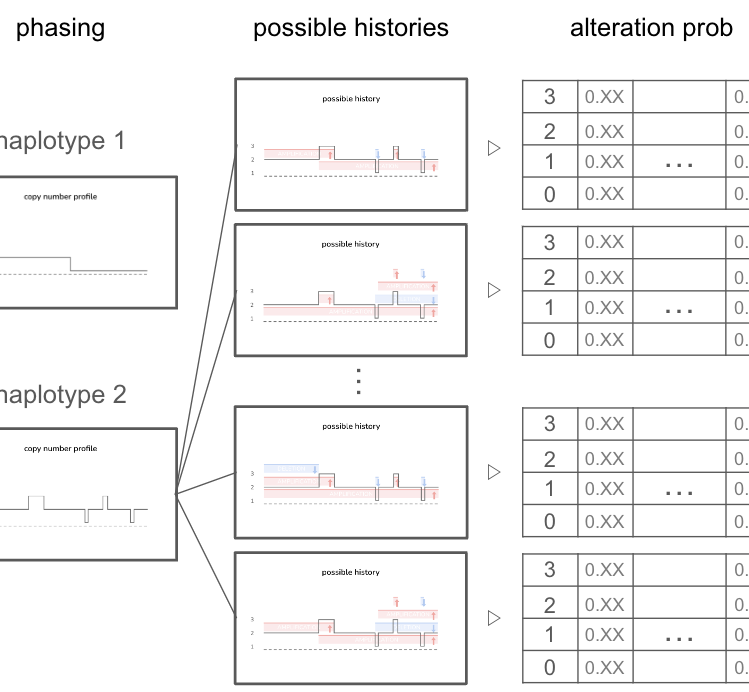
Cancer Algorithm Development
(2024)
Algorithmic Deconstruction of Somatic Copy Number Alterations for Therapeutic Target Discovery in Cancer
My research interest focuses on integrating computing/engineering methodologies to solve problems in biomedical sciences. I am particularly interested in developing and applying novel computational tools to understand how the complexities in the human genome contribute to diseases.

Algorithmic Deconstruction of Somatic Copy Number Alterations for Therapeutic Target Discovery in Cancer
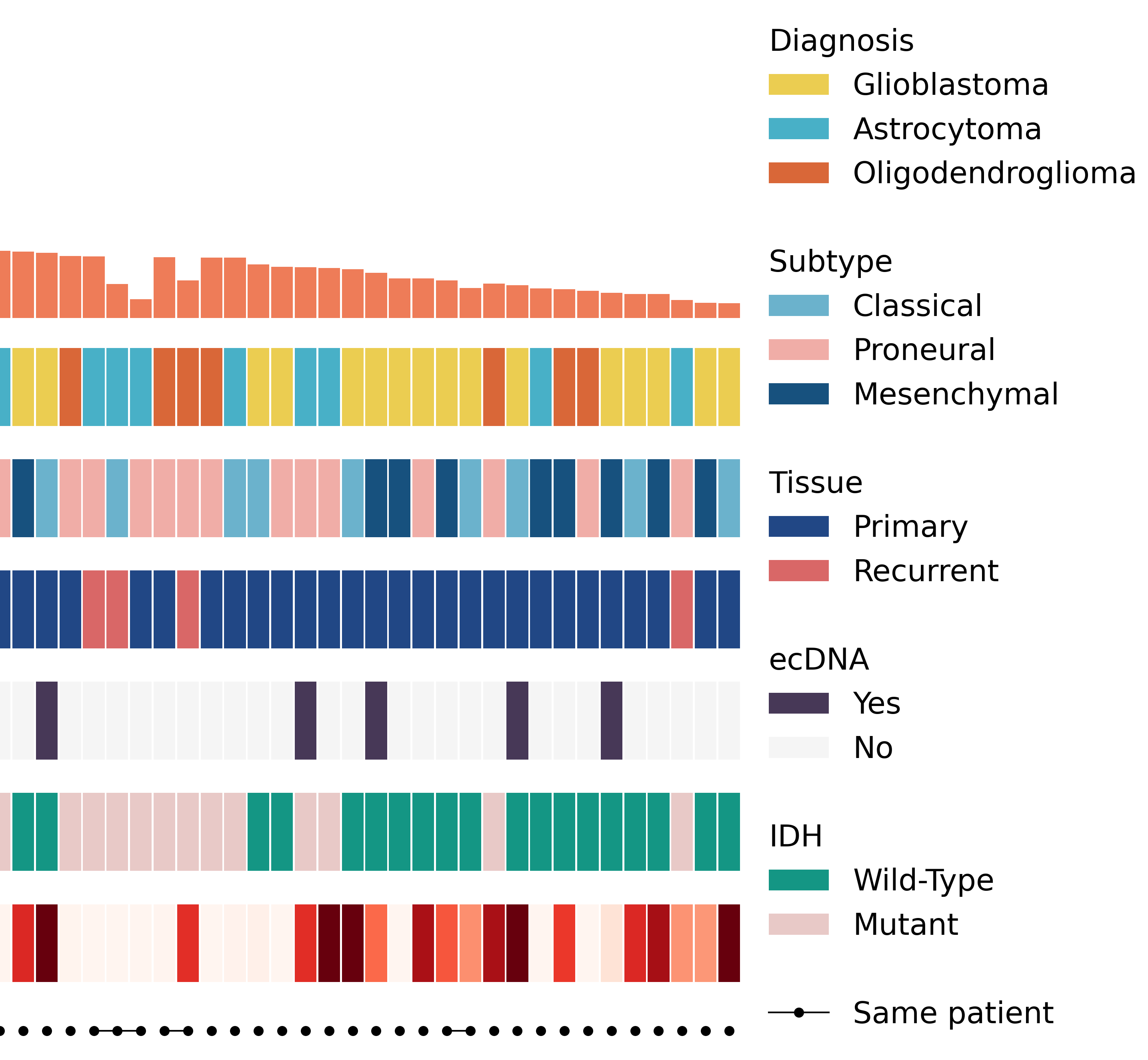
Structural Disruptions of the 3D Genome Architecture in Human Brain Cancer
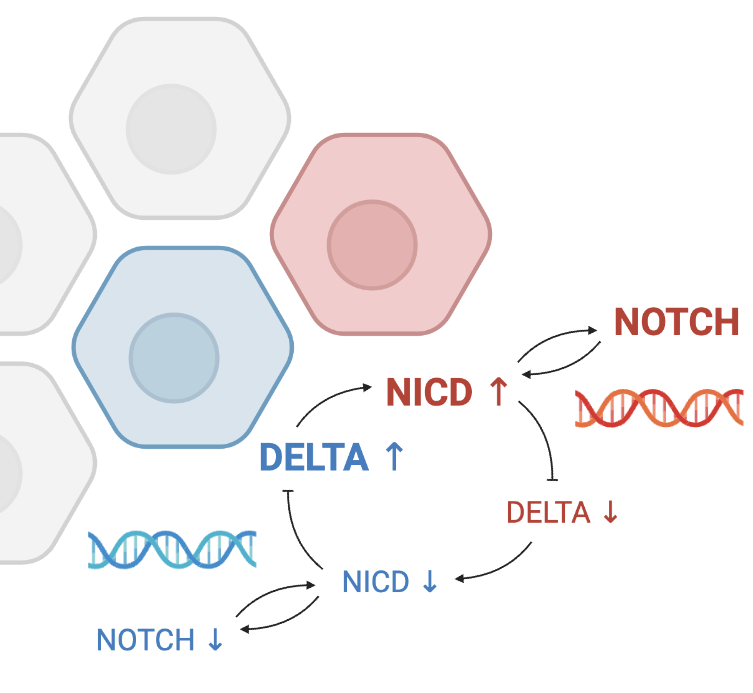
Investigating Cell Differentiation in the Brain with a Computational Model of Delta-Notch Signaling and Dynamical System Analysis
Modeling Perceptual Bistability: Impact of Neuronal Adaptation and Noise on Alternation Dynamics
Investigating the Efficacy of the Cascade Model of Synaptic Plasticity in a Biologically Constrained Simulation
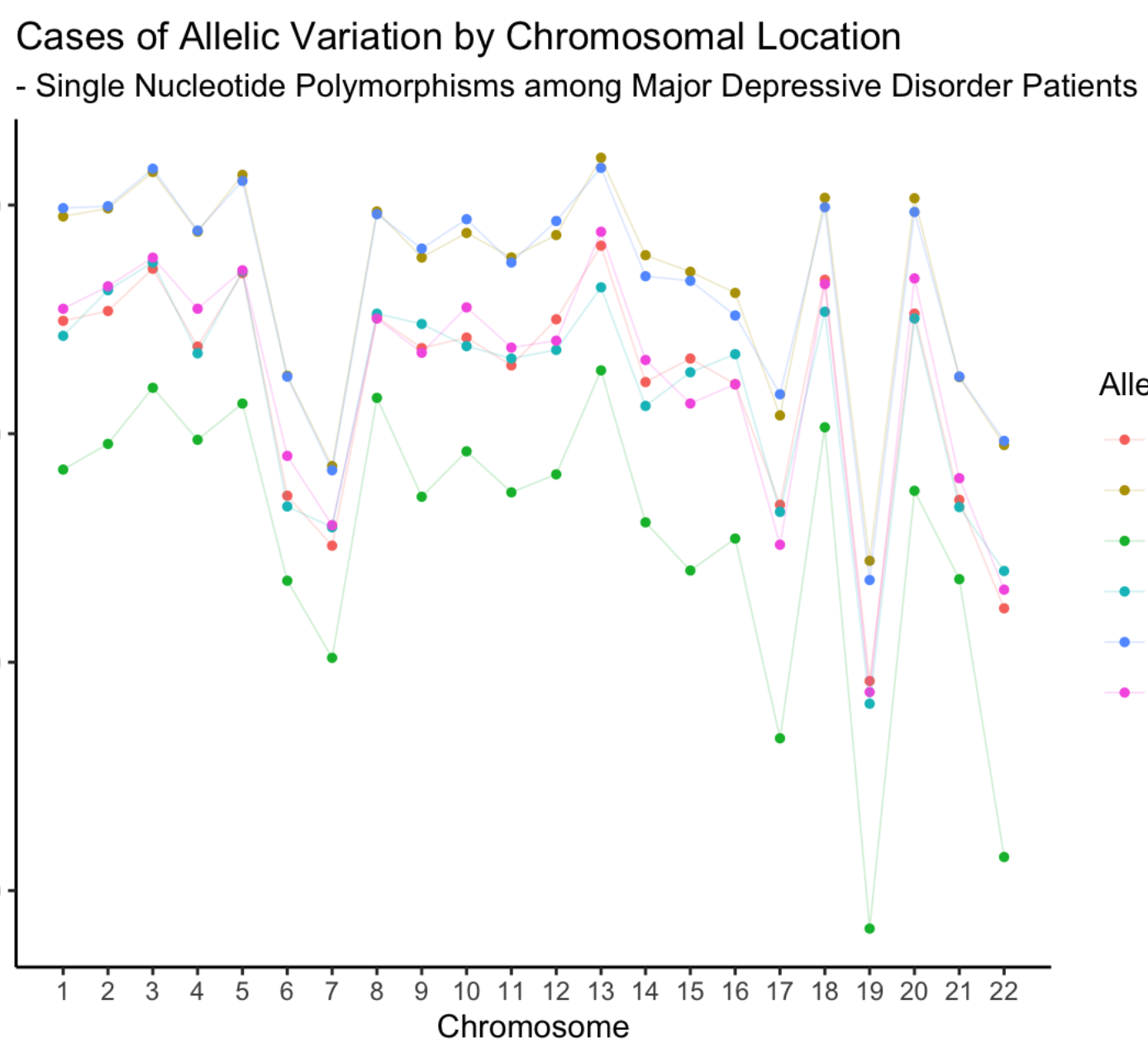
Comparative Analysis of Single Nucleotide Polymorphism (SNP) Genomic Data in Patients with Anxiety Disorder (AD) and Major Depressive Disorder (MDD)
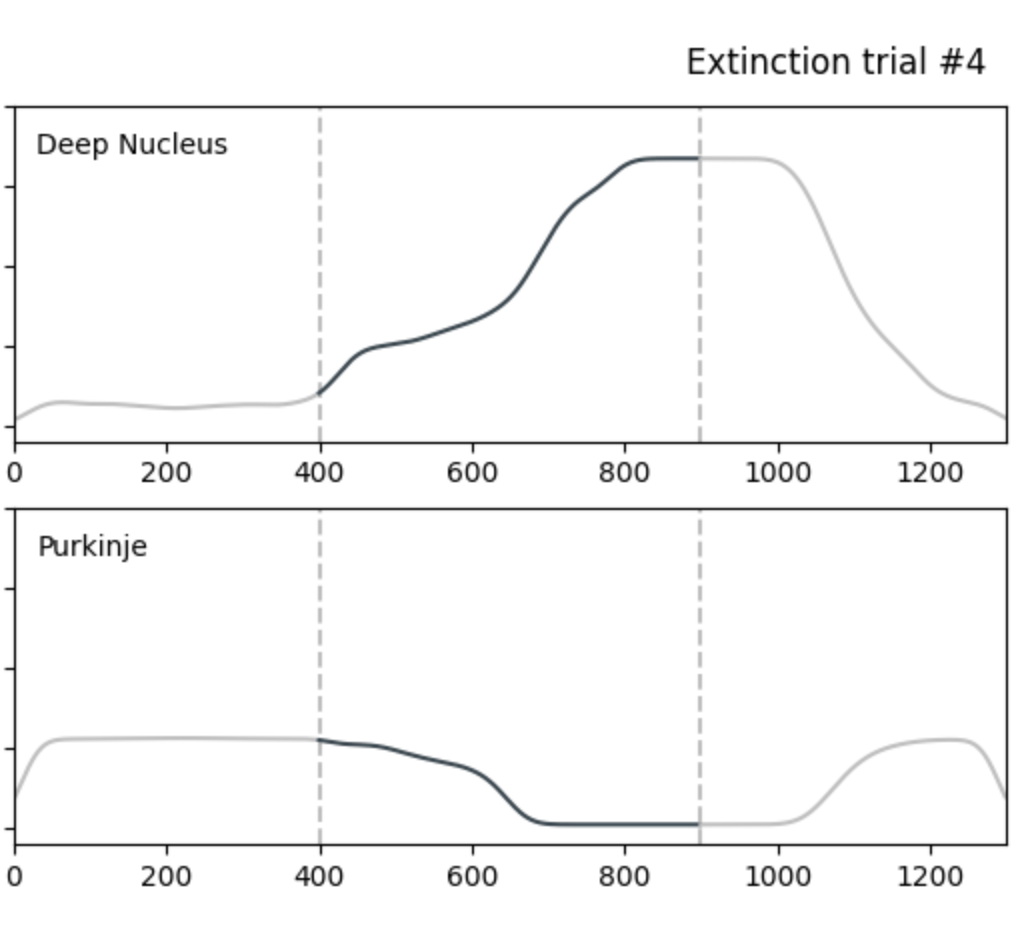
Simulation Analysis of Neuron Firing Activity in Cerebellar Cells for Learning and Extinction Evaluation
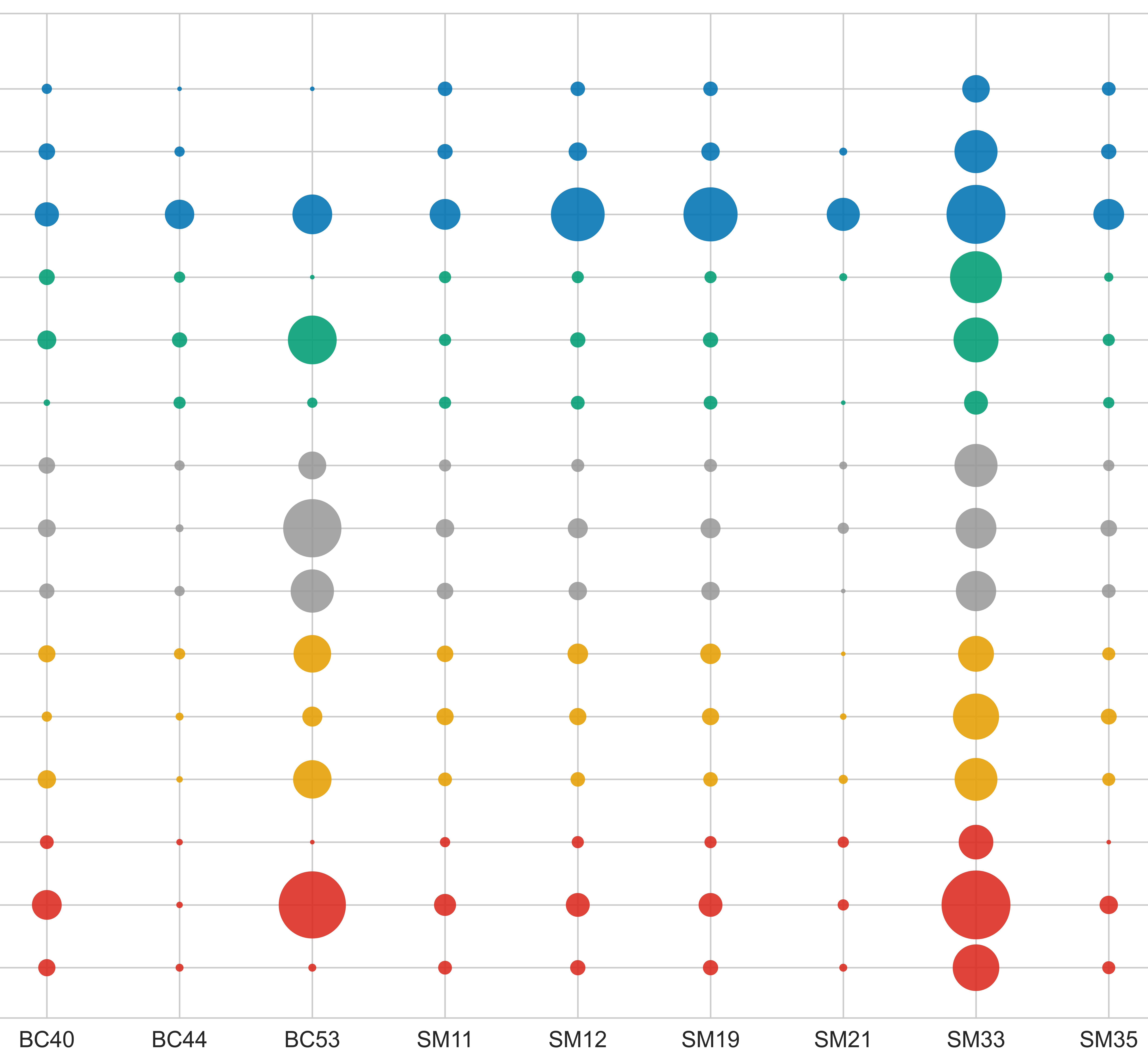
Computational Investigation of Single Nucleotide Driver Mutations and Tumor Evolution