Cancer 3D Genome Architecture
Structural Disruptions of the 3D Genome Architecture in Human Brain Cancer
This project was conducted during the Summer of 2023 in the Akdemir lab at the Department of Neurosurgery at MD Anderson Cancer Center.
Abstract
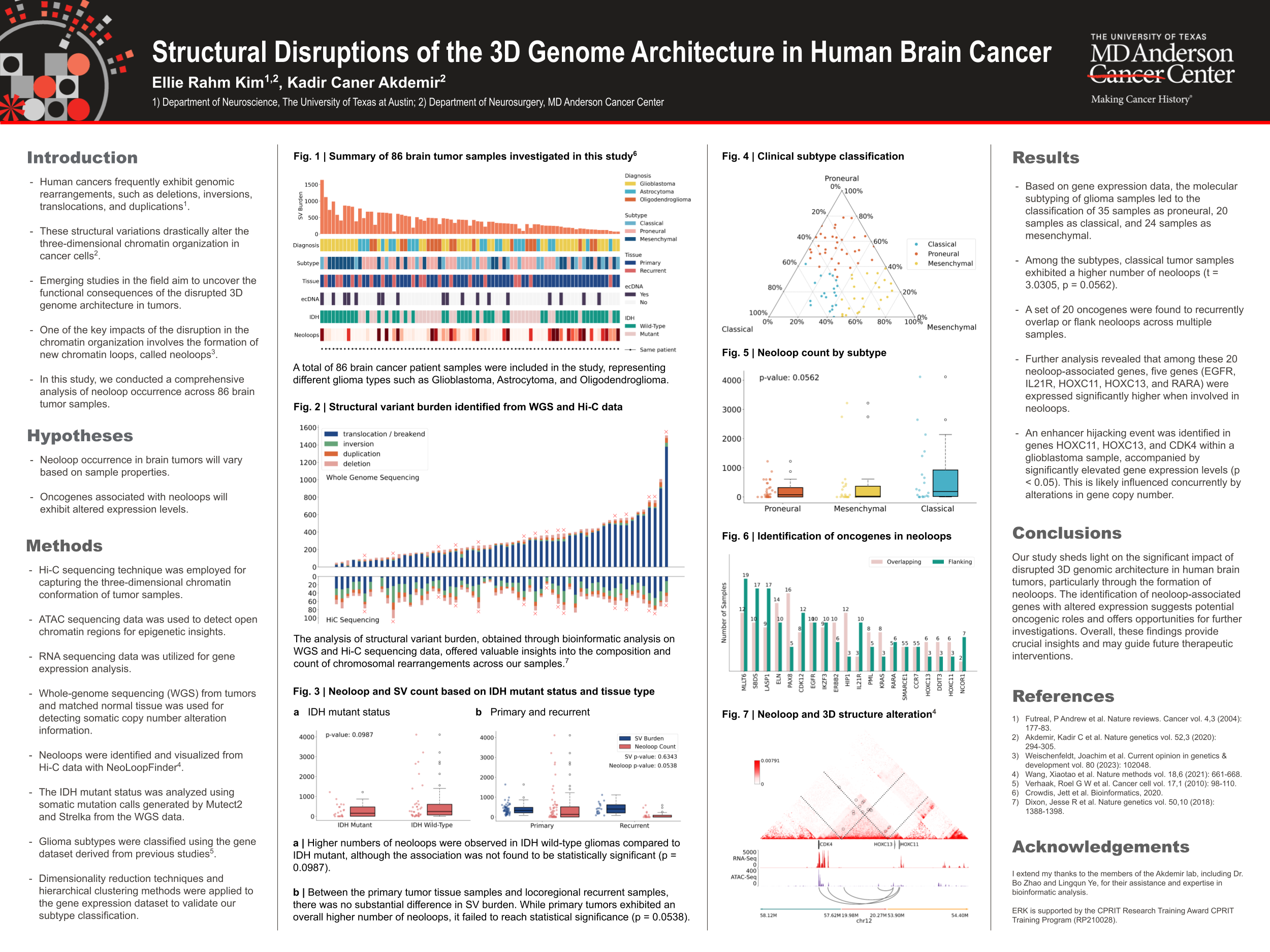
Genomic rearrangements in human cancers can lead to significant alterations in the three-dimensional (3D) chromatin organization, impacting gene expression and cancer development. The formation of new chromatin loops, termed neoloops, has emerged as a key consequence of disrupted 3D genome architecture in tumors. However, the occurrence of neoloops in brain cancer and their association with tumor properties, such as IDH mutant status, tissue type, and subtype, remain poorly understood. Additionally, the altered expression of neoloop-associated genes may play a significant role in driving oncogenic characteristics within tumor samples. To address these gaps, the objective of this research is to conduct a comprehensive analysis of neoloop occurrence across 86 brain tumor samples, providing valuable insights into the impact of disrupted 3D genomic architecture in brain cancer.
For this study, we performed an extensive analysis of neoloop occurrence in brain tumor samples using Hi-C sequencing data. We integrated Whole Genome Sequencing (WGS) data with five structural variant detection tools to identify copy number alterations and detect neoloops. RNA sequencing and ATAC sequencing data were utilized for gene expression and epigenetic analysis.
In this study, we analyzed a total of 86 patient samples, which represented various glioma types, including Glioblastoma, Astrocytoma, and Oligodendroglioma. Among the samples, IDH wild-type gliomas exhibited higher neoloop counts compared to IDH mutant gliomas, although not statistically significant (p = 0.0987). Additionally, classical glioma subtype samples displayed a higher number of neoloops compared to other subtypes (t = 3.0305, p = 0.0542). 20 recurrent neoloop-associated genes were identified, with five of them (EGFR, IL21R, HOXC11, HOXC13, and RARA) showing significantly elevated expression when involved in neoloops. Notably, an enhancer hijacking event in genes HOXC11, HOXC13, and CDK4 within a glioblastoma multiforme sample resulted in significantly elevated gene expression, presumably concurrently influenced by alterations in gene copy number.
Our study highlights the substantial impact of disrupted 3D genomic architecture in human brain tumors, particularly through neoloop formation. The identification of neoloop-associated genes with altered expression suggests potential oncogenic roles and offers opportunities for further investigations in cancer biology. These findings provide crucial insights into cancer initiation and progression and may guide future therapeutic interventions.